In a groundbreaking study published in Nature Methods, researchers unveiled a novel and highly sensitive proteomics method called the peptide-centric local stability assay (PELSA). This innovative technique enables the simultaneous identification of ligand-binding proteins and their binding sites in complex biological systems. It represents a significant advancement in the field of proteomics, offering broad applicability to diverse ligands, including metabolites, pharmaceuticals, and environmental pollutants.
Proteins are at the heart of virtually all biological processes, and their functions often depend on interactions with ligands—molecules that bind to specific sites on proteins. Ligands play a wide range of roles, acting as enzyme substrates or inhibitors, signaling molecules, allosteric modulators, or structural stabilizers. Studying protein-ligand interactions is therefore essential for several key areas of research. These include understanding the functions of uncharacterized proteins, uncovering regulatory mechanisms in cellular metabolism, elucidating the mechanisms of drug action, and designing drugs through structure-based approaches. Furthermore, identifying the specific regions on proteins where ligands bind provides valuable insights for advancing biological hypotheses and therapeutic strategies.
Conventional methods for investigating ligand-binding sites and measuring binding affinities often require the purification of recombinant proteins. While effective, this approach is labor-intensive, time-consuming, and can fail to replicate the proteins’ native cellular environments. These limitations may lead to inaccuracies in assessing binding affinities and protein behavior under physiological conditions.
Recent developments in modification-based proteomics methods have provided a promising alternative. These methods identify ligand-binding proteins and their interaction sites directly within native cellular lysates, bypassing the need for protein purification. However, many of these approaches require chemical modifications to the ligand, which can interfere with its natural activity or render certain ligands unsuitable for analysis. This is particularly problematic for ligands that are structurally diverse or exhibit low binding affinities, such as many metabolites.
To address these challenges, researchers led by Professor Ye Mingliang from the Dalian Institute of Chemical Physics of the Chinese Academy of Sciences (CAS), in collaboration with Professor Luo Cheng’s group from the Shanghai Institute of Materia Medica of CAS, developed PELSA. This method utilizes an innovative approach to assess protein stability and ligand interactions in their native states.
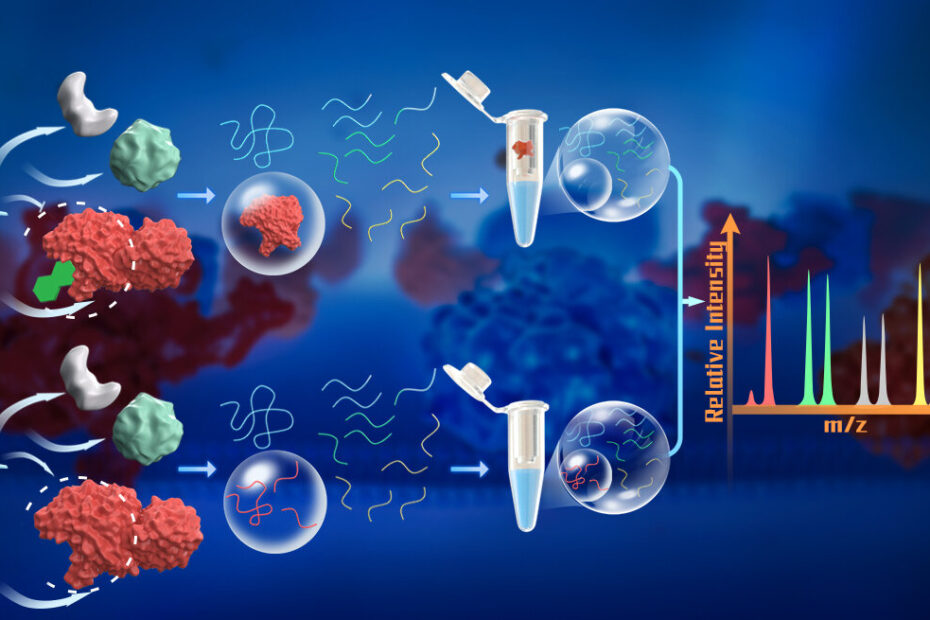
The PELSA technique employs a large quantity of trypsin, a proteolytic enzyme, to generate small peptides directly from native proteins. By using a high enzyme-to-substrate (E/S) ratio of 1:2, the method ensures that even protein regions in low-energy states are cleaved, producing peptides that accurately reflect the local stability of the parent proteins. Importantly, this process is performed under native conditions, preserving the proteins’ physiological states, including post-translational modifications and interactions with other cellular components.
The generated peptides are separated from partially digested proteins and analyzed using mass spectrometry. By comparing peptide abundance between ligand-treated and untreated samples, researchers can identify the ligand-binding regions of proteins and the specific proteins involved. This method eliminates the need for ligand chemical modification, making it applicable to a broader range of ligands.
PELSA has demonstrated remarkable sensitivity and effectiveness in target protein identification. For example, when applied to the pan-kinase inhibitor staurosporine, PELSA achieved a 12-fold increase in kinase target identification compared to LiP-MS, a state-of-the-art modification-free proteomics method. Additionally, PELSA identified 2.4 times more kinase targets than thermal proteome profiling (TPP), a widely used technique that lacks the ability to provide detailed binding site information. PELSA’s dose-dependent experimental design also enables the measurement of local binding affinities, offering deeper insights into dynamic structural changes in proteins upon ligand binding under physiological conditions.
One of the significant strengths of PELSA is its ability to tackle the challenges posed by metabolites. Metabolites, known for their structural diversity and low binding affinities, have historically been difficult to study using traditional methods. PELSA has proven particularly effective in systematically identifying metabolite-binding proteins. For instance, in experiments involving alpha-ketoglutarate in HeLa cell lysates, PELSA identified 40 candidate target proteins, 30 of which were already established as alpha-ketoglutarate binding proteins. This result highlights the method’s sensitivity and reliability. PELSA also successfully identified binding proteins for other metabolites, such as folate, leucine, fumarate, and succinate, further demonstrating its broad applicability.
PELSA offers several advantages over existing techniques. Unlike methods that require ligand modification or rely on purified proteins, PELSA can directly detect ligand-induced changes in protein stability within total cell lysates. This allows researchers to analyze proteins in their native, physiologically relevant states, which include natural post-translational modifications and protein-protein interactions. The method’s ability to provide detailed information about ligand-binding sites and local binding affinities makes it an invaluable tool for structural biology, drug discovery, and systems biology research.
This advancement represents a significant step forward in the field of proteomics. By enabling a more comprehensive and accurate analysis of protein-ligand interactions, PELSA has the potential to transform our understanding of biochemical processes and accelerate the development of novel therapeutic strategies. With its broad applicability, high sensitivity, and ability to preserve the native state of proteins, PELSA is poised to become a cornerstone method for studying protein-ligand interactions in complex biological systems.
Source: Chinese Academy of Sciences